Case Study: Magnaporthe oryzae
The experimentally validated nucleolar proteins from M. oryzae and their prediction scores in the FuNGI.
Background
Magnaporthe oryzae, a filamentous fungus responsible for rice blast disease, is a well-established model organism for plant–pathogen interaction studies. With its genetic tractability and extensive experimental background, it serves as an ideal system to evaluate the predictive performance of FuNGI. As a representative filamentous ascomycete with a complex lifecycle and specialized infection structures (such as appressoria), M. oryzae offers unique insights into fungal cell biology that go beyond what can be studied in yeasts.
To date, most limited knowledge of fungal nucleolar proteins has been derived from the model yeast Saccharomyces cerevisiae, which possesses a simplified bipartite nucleolus. However, this model may not fully reflect the nucleolar complexity found across the fungal kingdom. Especially, filamentous fungi—given their complex developmental and pathogenic programs—are expected to diverge even further. Yet, nucleolus in filamentous fungi remains largely unexplored. This highlights the importance of expanding systematic nucleolar studies to filamentous fungus like M. oryzae, in order to better understand the evolutionary and functional diversity of nucleoli across fungi.
Fluorescence Microscopy
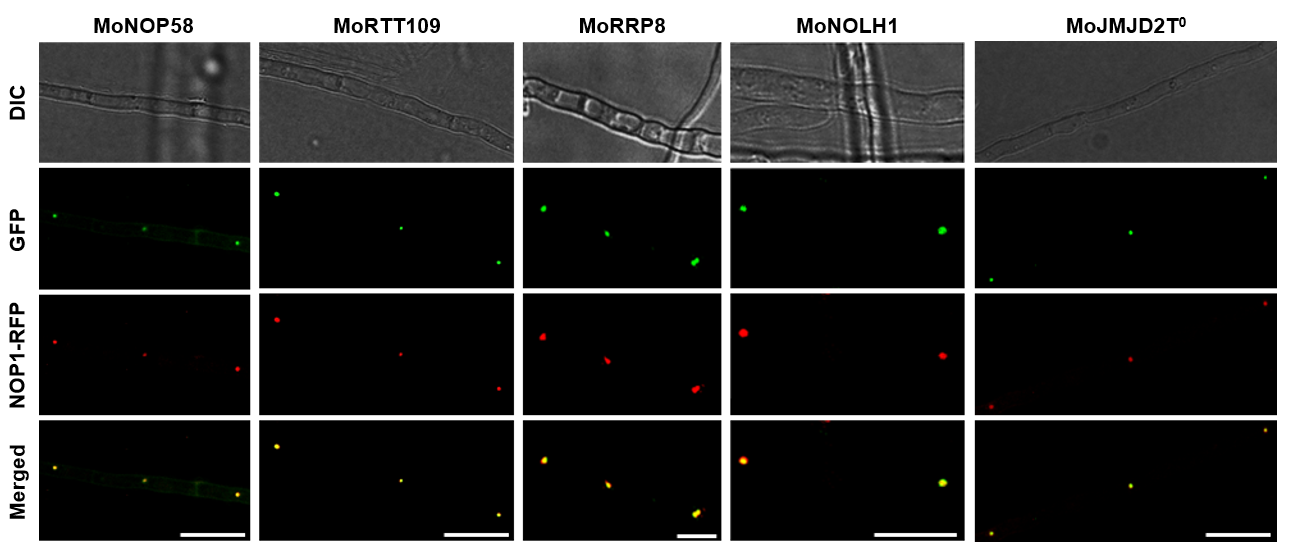
Figure 1. Co-localization of MoNop1-RFP with GFP-tagged candidate nucleolar proteins predicted by FuNGI in Magnaporthe oryzae hyphae.
Fluorescence microscopy images of M. oryzae hyphae co-expressing the nucleolar marker MoNop1-RFP and individual GFP-tagged candidate proteins predicted to localize to the nucleolus by FuNGI. From left to right: representative images of hyphae expressing MoNop1-RFP with MoNOP58-GFP, MoRTT109-GFP, MoRRP8-GFP, MoNOLH1-GFP, and MoJMJD2T0-GFP. For each strain, differential interference contrast (DIC), green fluorescence (GFP), red fluorescence (MoNop1-RFP), and merged images are shown. Yellow puncta in the merged panels indicate co-localization of candidate proteins with the nucleolar marker. Scale bars = 10 µm.
Experimental Design & Result
To validate the predictions of FuNGI, five NoLS-positive proteins from M. oryzae were randomly selected, including four with high-confidence scores and one with a medium-confidence score (Table 1).
To generate fluorescent fusion constructs, the coding sequences (excluding stop codons) of MoNOP58, MoRTT109, MoRRP8, MoNOLH1, and MoJMJD2T0 were amplified from M. oryzae KJ201 cDNA and fused in-frame to the N-terminus of green fluorescent protein (GFP) under the control of their predicted native promoter using the double-joint PCR strategy (Yu et al., 2004). Each GFP fusion construct was cloned into a T&A cloning vector (RBC, Taipei, Taiwan) and subsequently transformed into the transgenic M. oryzae strain expressing MoNOP1-RFP, a nuclear marker. Transformants were grown on sterile slide glasses coated with CM agar at 25ºC for 5 days.
Differential interference contrast (DIC) and fluorescence microscopy were performed using a Leica DM2500 light microscope (Leica, Wetzlar, Germany), and images were acquired with a Leica DFC7000 T digital camera (Leica, Wetzlar, Germany). Fluorescence excitation was set at 480/40 nm for GFP and 515–560 nm for RFP. Images were processed using LAS X software (Leica, Wetzlar, Germany).
Fluorescence microscopy analysis revealed that all five candidate proteins, as predicted by FuNGI, localized to the nucleolus in M. oryzae hyphae. Each GFP-tagged protein (green) displayed a strong co-localization with the nucleolar marker MoNop1-RFP (red) in the nuclei, as confirmed by the overlapping fluorescence signals (Figure 1). These results experimentally validate the nucleolar localization of the candidate proteins.
Prediction Scores
Their prediction features of the five candidate proteins are summarized below:
| Locus | Symbol | Accession | Protein name | Length | #NoLS | #NLS | PSORT | Score | Confidence |
|---|---|---|---|---|---|---|---|---|---|
| MGG_00053 | MoNOLH1 | G4NET3 | NUC153 domain-containing protein | 728 | 5 | 5 | Nuclear | 22.6 | High |
| MGG_05969 | MoRTT109 | G4N4I8 | Histone acetyltransferase | 579 | 1 | 1 | Nuclear | 12.1 | Medium |
| MGG_07008 | MoNOP58 | G4MPA1 | Nucleolar protein 58 | 599 | 3 | 2 | Cytoplasm | 16.6 | High |
| MGG_07132 | MoRRP8 | G4MT55 | Ribosomal RNA-processing protein 8 | 507 | 3 | 6 | Cytoplasm | 18.2 | High |
| MGG_09186 | MoJMJD2 | G4MPD2 | [Histone H3]-trimethyl-L-lysine(9) demethylase | 1529 | 2 | 3 | Nuclear | 22.4 | High |
Table 1. List of Magnaporthe oryzae proteins predicted with NoLS and NLS used for experimental validation
Secondary Structures of NoLS regions
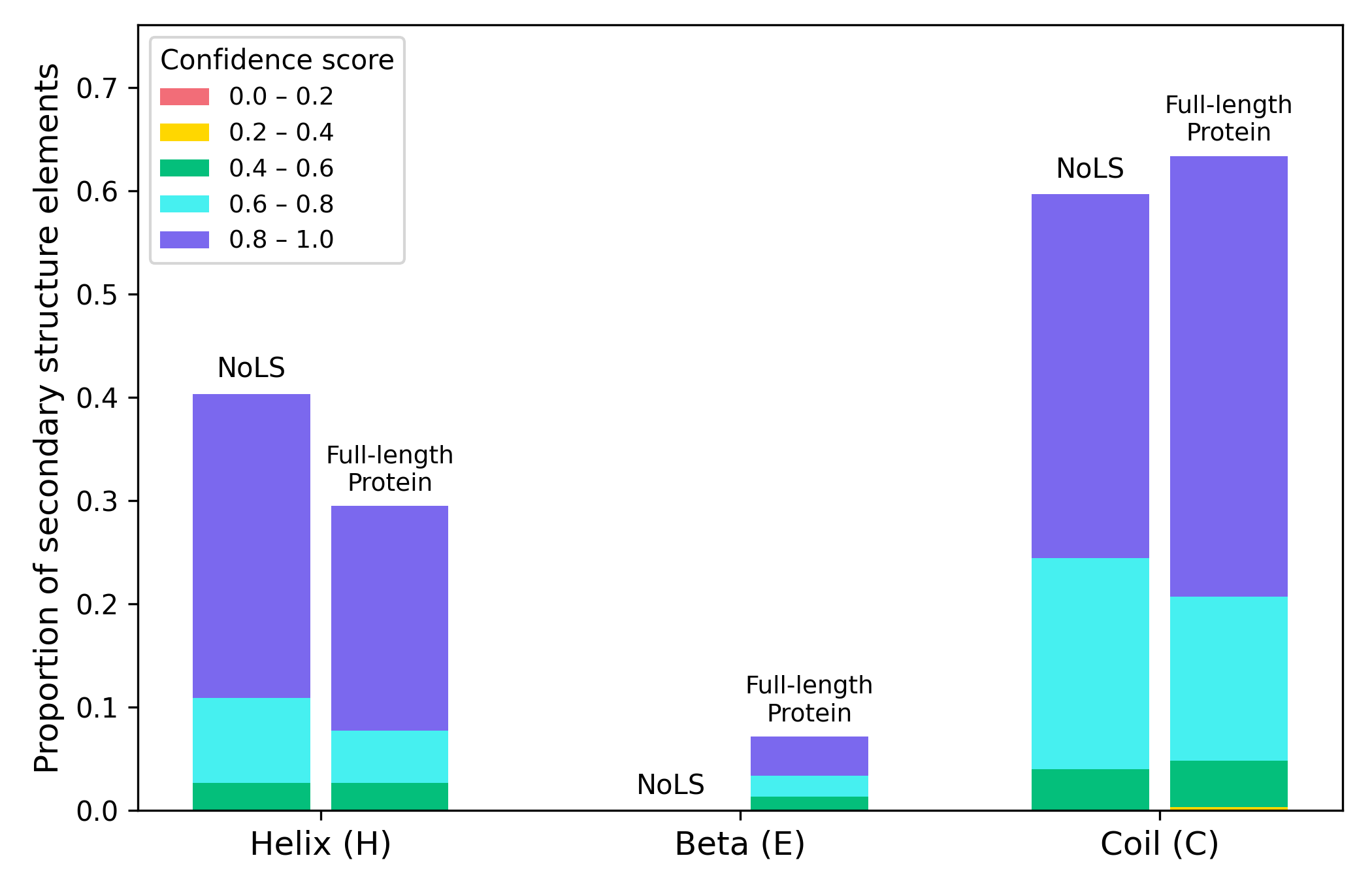
Figure 2. Proportion of secondary structure elements within NoLS regions and corresponding full-length sequences of five experimentally validated M. oryzae proteins.
PSIPRED predictions (Buchan et al., 2019) are grouped by structural class (α-helix, β-strand, coil) and subdivided into confidence score intervals ([0.0–0.2] to [0.8–1.0]). NoLS regions occur predominantly within α-helices or coils, with β-strands not observed.
Conclusion
The successful localization of all five proteins to the nucleolus strongly supports the predictive validity of FuNGI. These results demonstrate that the integrated scoring framework of FuNGI can effectively prioritize candidate nucleolar proteins for experimental characterization.
Reference
Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Domínguez Y, Scazzocchio C. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004. 41(11):973–81.
Buchan DWA, Jones DT. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 2019. 47(W1):W402–7.